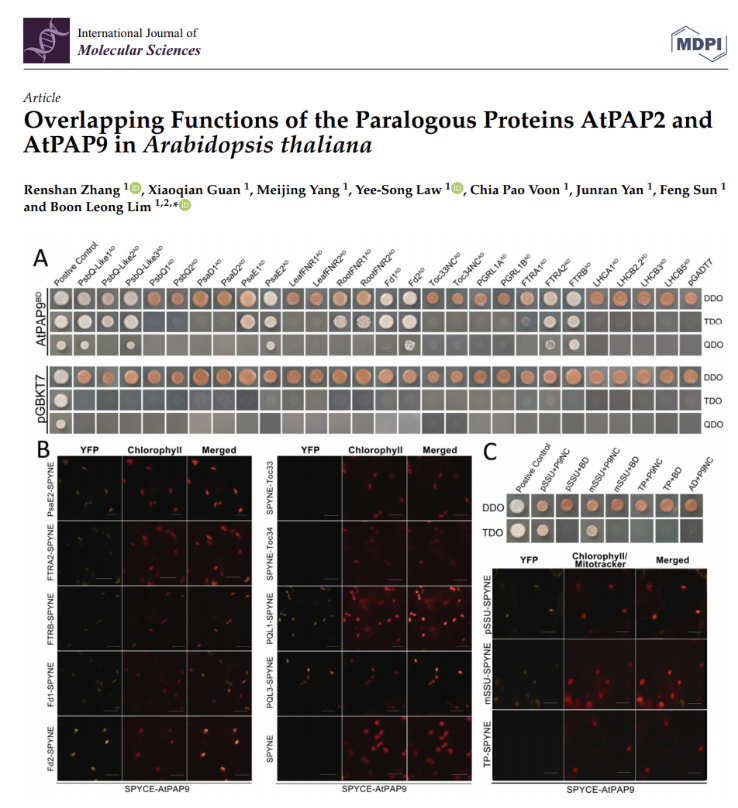
Overlapping Functions of the Paralogous Proteins AtPAP2 and AtPAP9 in Arabidopsis thaliana
Arabidopsis thaliana purple acid phosphatase 9 (AtPAP9) displays a 72% sequence similarity with AtPAP2, and both phosphatases carry hydrophobic C-terminal motifs. Prof BL Lim’s team showed that AtPAP9 and AtPAP2 serve partially redundant biological functions in chloroplast protein import.
Full Version
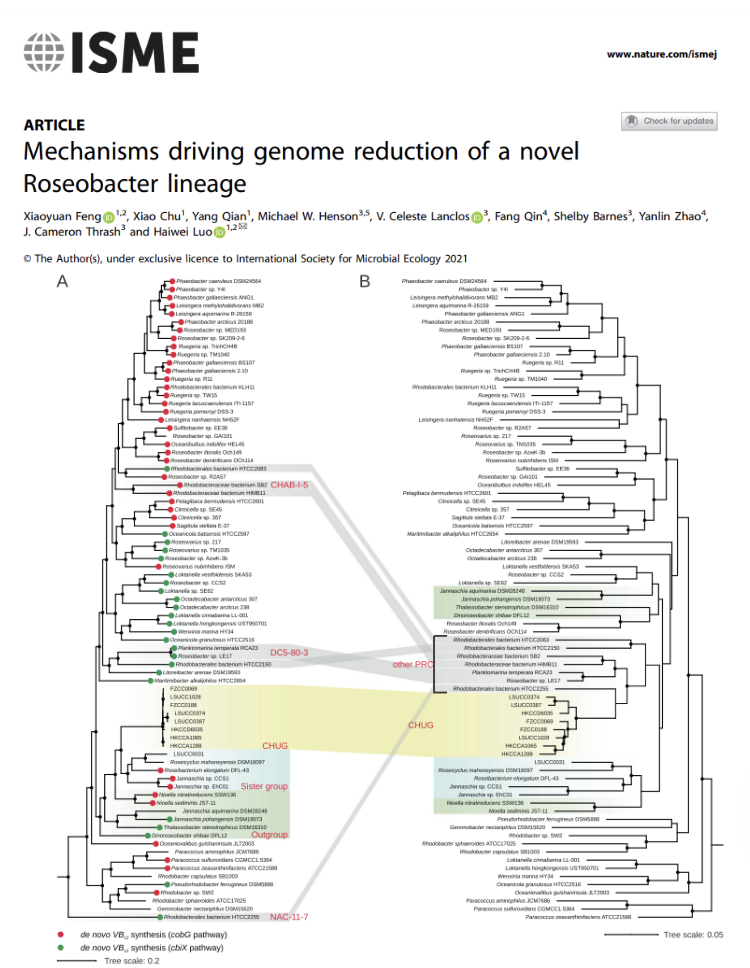
Mechanisms driving genome reduction of a novel
Roseobacter lineage
Prof HW Luo’s team discovered a novel Alphaproteobacteria lineage very abundant in the environment. Members of this lineage have small genomes and cannot synthesize vitamin B12, making them distinctly different from all their sister lineages. Further analyses suggest that they may interact with aquatic plants in a way different from their sister lineages.
Full Version
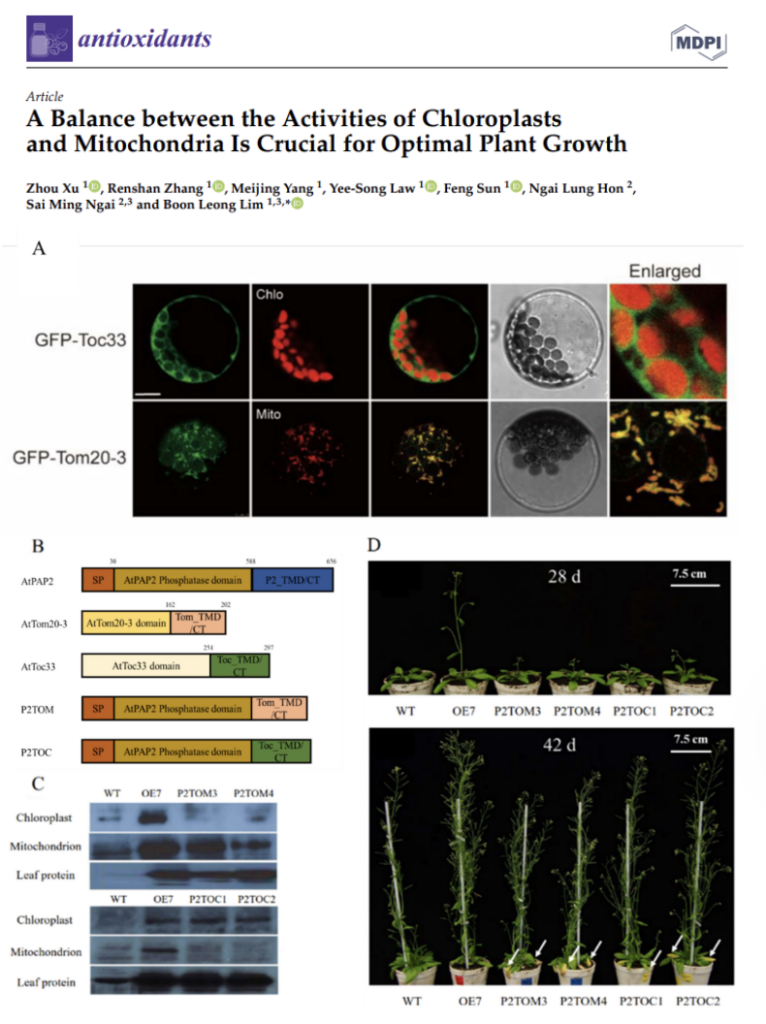
A Balance between the Activities of Chloroplasts
and Mitochondria Is Crucial for Optimal Plant Growth
Prof BL Lim’s team showed that overexpression of Arabidopsis thaliana purple acid phosphatase 2 (AtPAP2) solely in chloroplasts or mitochondria boost their activities respectively. These transgenic lines can be useful tools for studying how hyperactive chloroplasts or mitochondria affect cellular metabolism and plant physiology.
Full Version
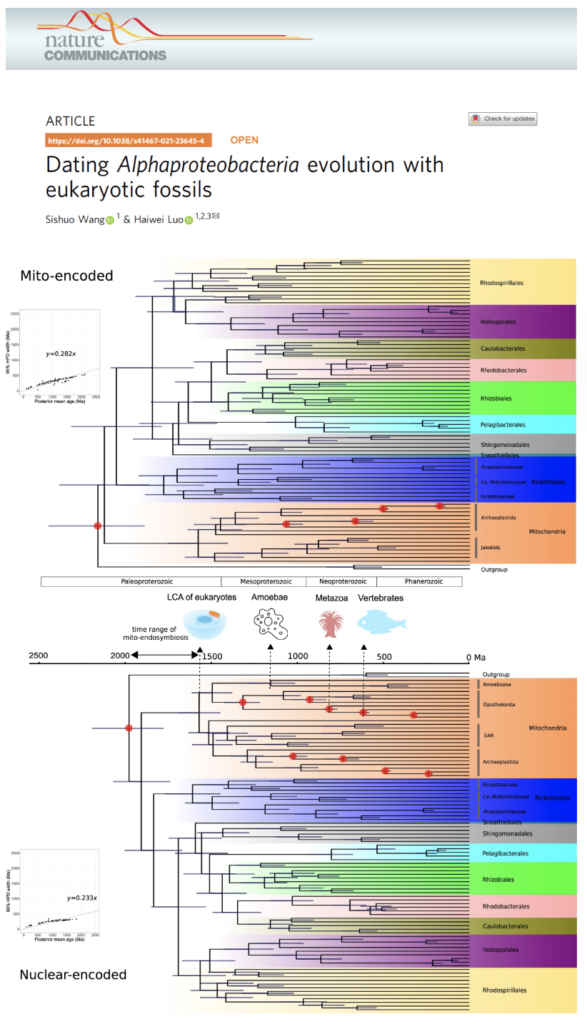
Dating Alphaproteobacteria evolution with eukaryotic fossils
Prof HW Luo’s team incorporated eukaryotic fossils to date the divergence times of Alphaproteobacteria, based on the mitochondrial endosymbiosis that mitochondria evolved from an alphaproteobacterial lineage.
Full Version
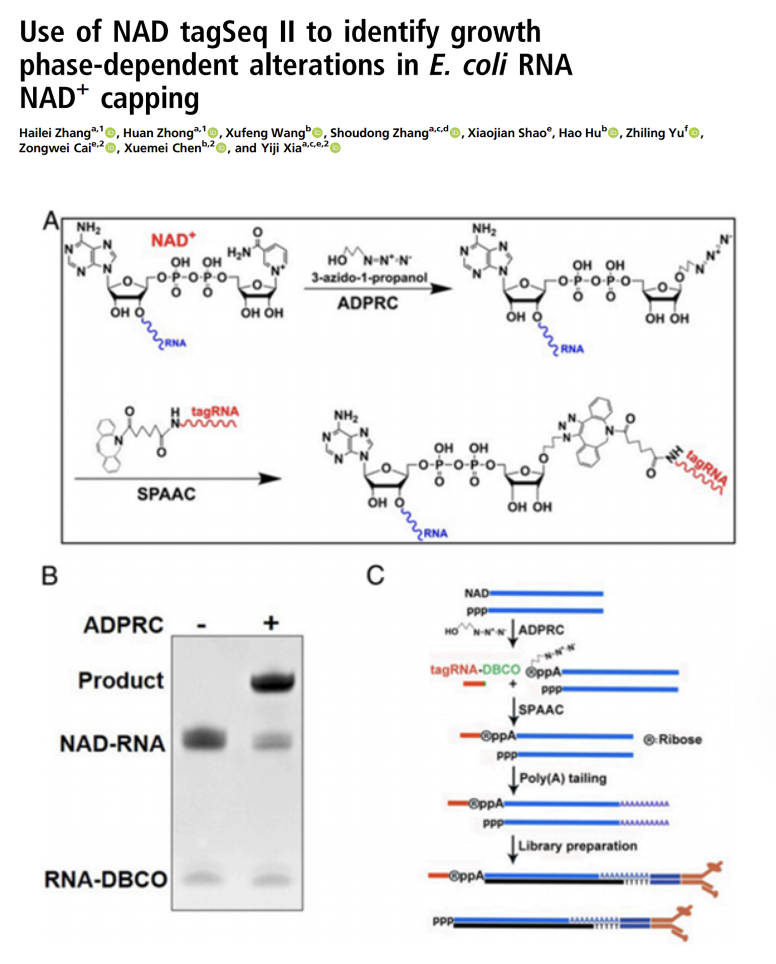
Use of NAD tagSeq II to identify growth phase-dependent alterations in E. coli RNA NAD+ capping
The development of NAD tagSeqII to compare NAD-RNA and total transcript profile of E. coli cells in the exponential and stationary phases.Related article: SPAAC-NAD-seq, a sensitive and accurate method to profile NAD+-capped transcripts https://www.pnas.org/content/118/13/e2025595118.long
Full Version
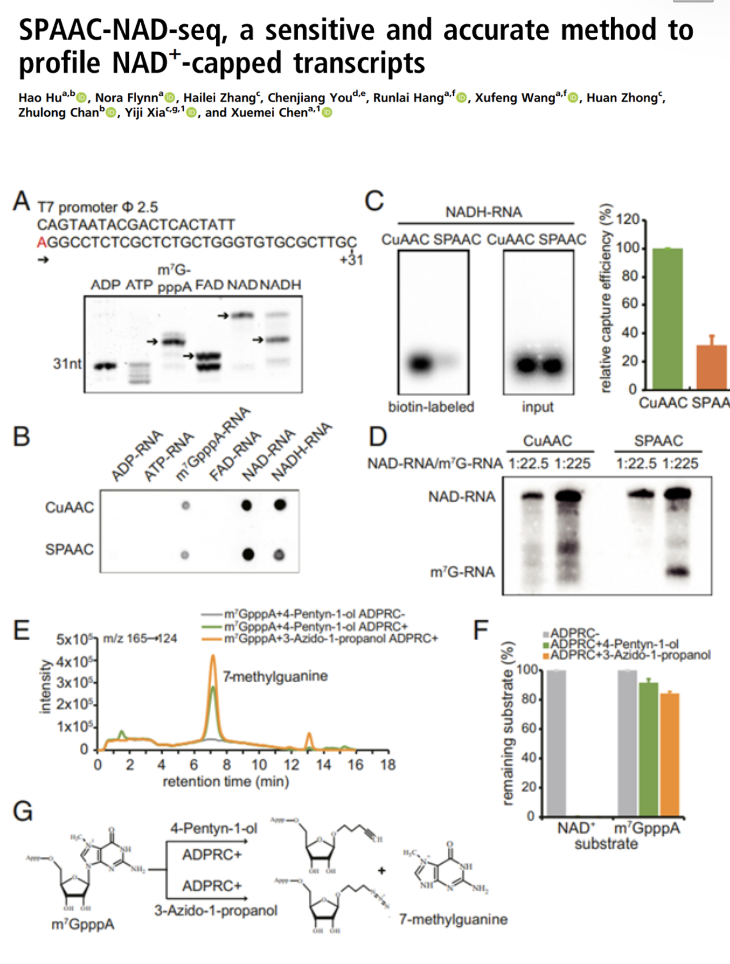
SPAAC-NAD-seq, a sensitive and accurate method to profile NAD+-capped transcripts
Prof Xia’s team has developed a new method to sequence NAD+-capped transcripts. Compared with the old methods, this new method showed higher capturing efficiency, higher specificity and is able to maintain higher integrity.
Full Version
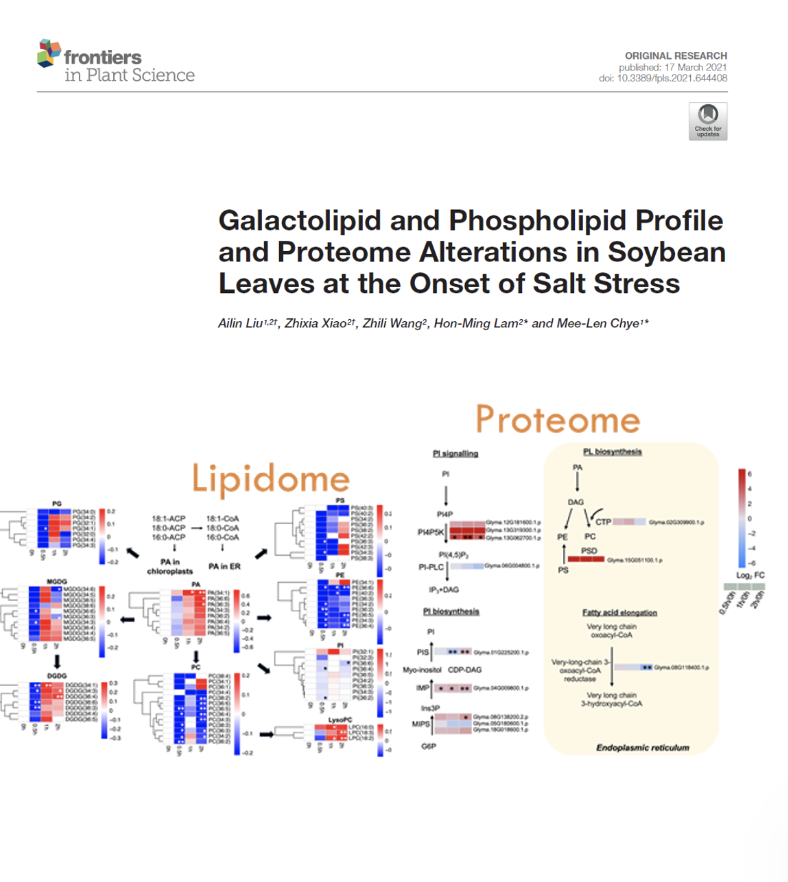
Galactolipid and Phospholipid Profile and Proteome Alterations in Soybean Leaves at the Onset of Salt Stress
Prof Chye and Prof Lam have teamed up to explore the adaptive mechanisms underpinning lipid homeostasis of soybean leaves at the on set of salt stress, lipid profiling and proteomic analyses were performed. Our results demonstrated that salt stress resulted in a remodelling of membrane lipid composition and an alternation in membrane lipids associated with lipid signalling and metabolism.
Full Version
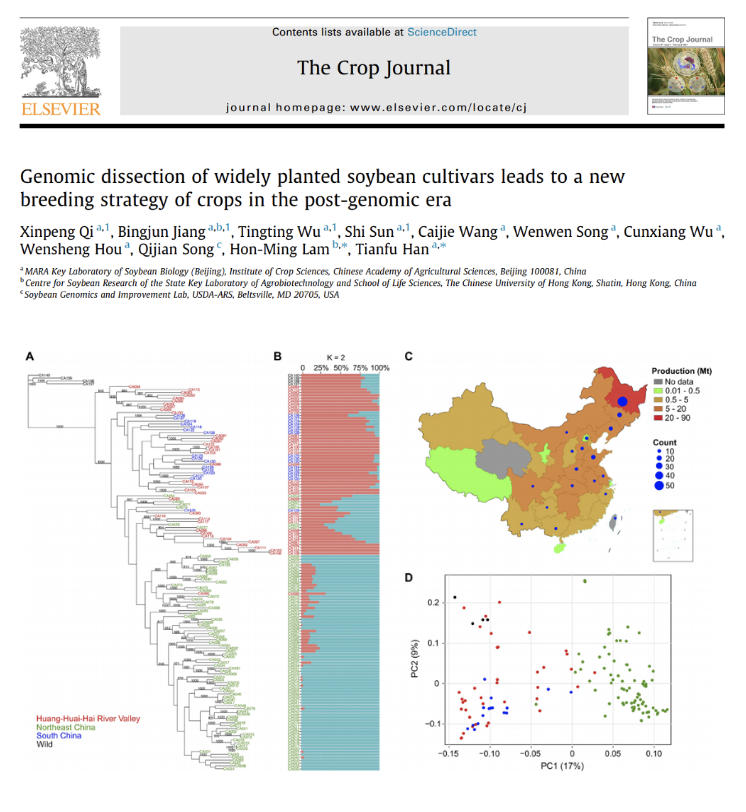
Genomic dissection of widely planted soybean cultivars leads to a new breeding strategy of crops in the post-genomic era
In collaboration with Prof Tianfu Han in the Chinese Academy of Agricultural Sciences, the center has participated in the genomic analysis of widely planted soybean cultivars in the past 100 years.
Full Version
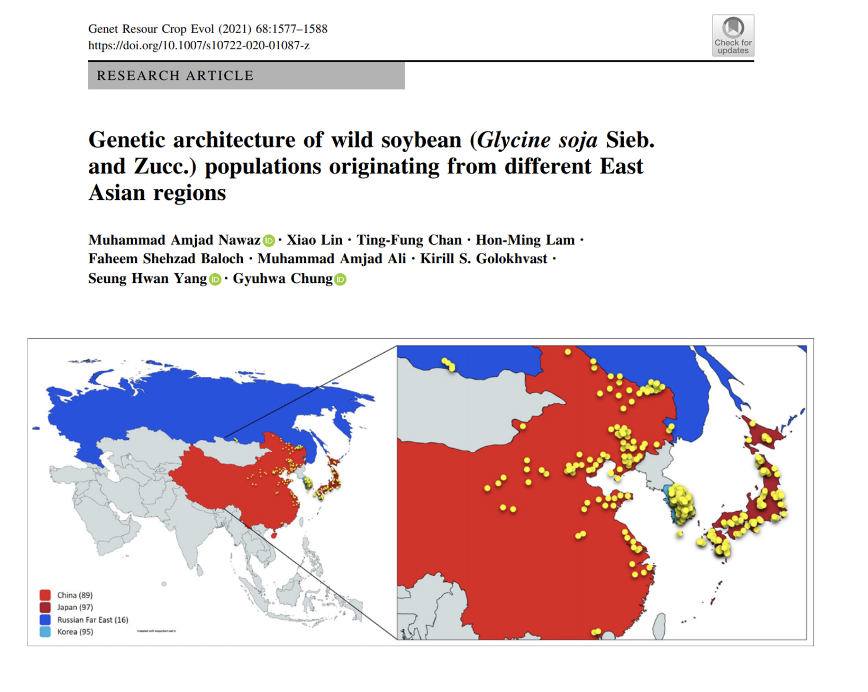
Genetic architecture of wild soybean (Glycine soja Sieb. and Zucc.) populations originating from different East Asian regions
In collaboration with Prof Gyuhwa Chung in Chonnam National University, South Korea, the AoE has analysed the genetic structure of the wild soybean populations from four East-Asian geographies including China, Japan, Korea, and Russian Far East.
Full Version
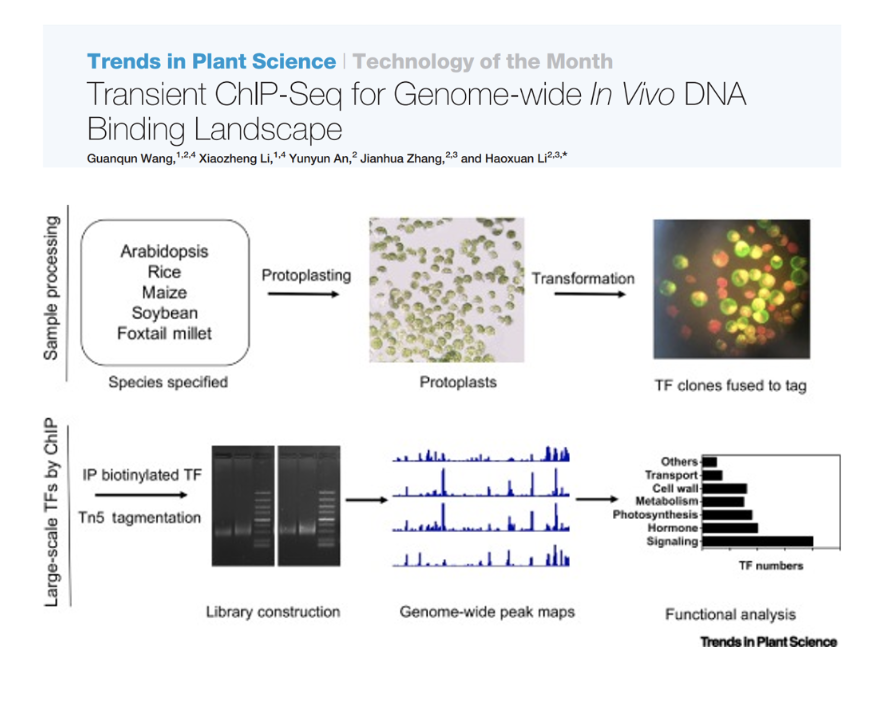
Transient ChIP-Seq for Genome-wide In Vivo DNA Binding Landscape
Technology of the Month. We have introduced the transient ChIP-Seq in the Trends in Plant Science.
Full Version
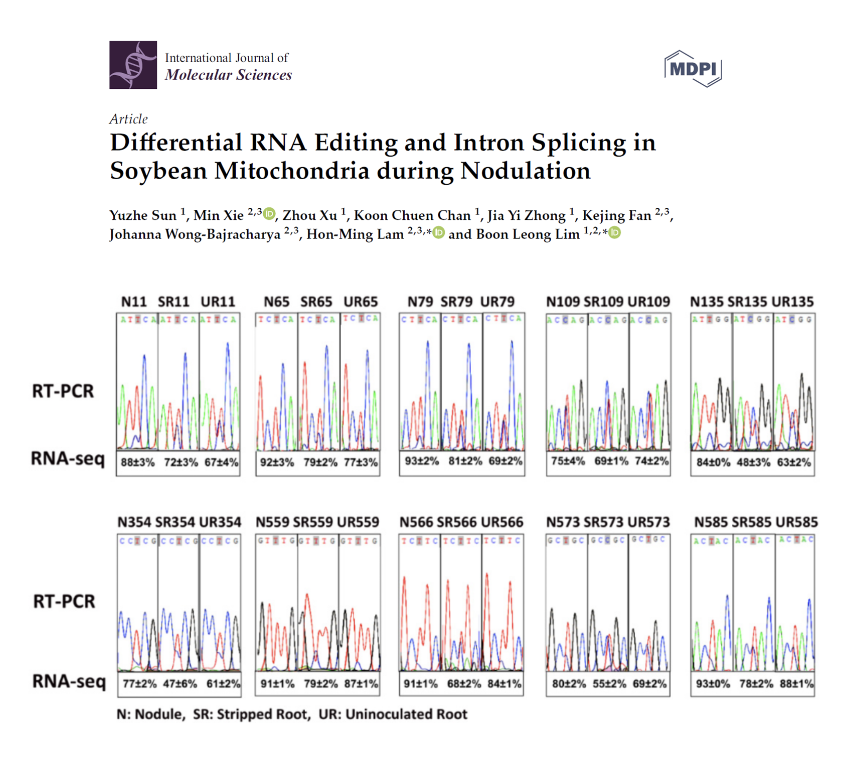
Differential RNA Editing and Intron Splicing in Soybean Mitochondria during Nodulation
Alternation of mitochondrial proteome and transcriptome could have contributed to the change in energy status during nodulation and nitrogen fixation. Through RNA-seq, differentially edited sites on mitochondrial transcripts were identified. Eight out of these sites were located on the matR transcripts which encode the maturase for transcript splicing. At the same time, the splicing efficiencies of a few mitochondrial transcripts were also found altered in nodules.
Full Version
Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors
Prof Silin Zhong’s team has performed a large scale ChIP-seq project to reconstruct a transcription network of 104 transcription factors in maize leaf.
Full Version
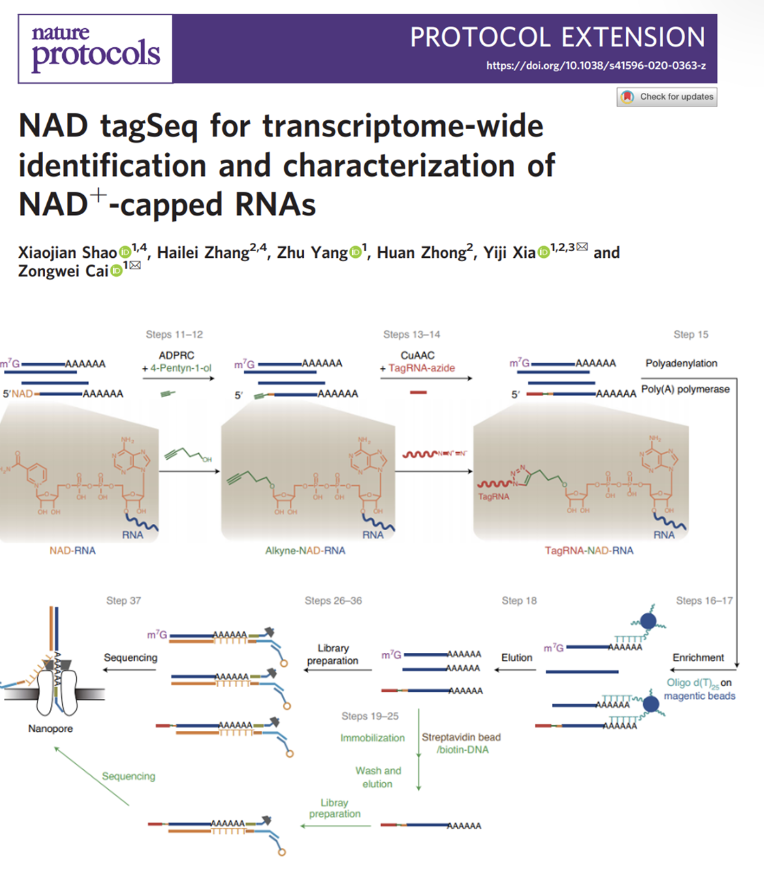
NAD tagSeq for transcriptome-wide identification and characterization of NAD+-capped RNAs
A detail protocol of the NAD tagSeq was published in Nature Protocols. Link to original research: https://www.pnas.org/content/116/24/12072
Full Version
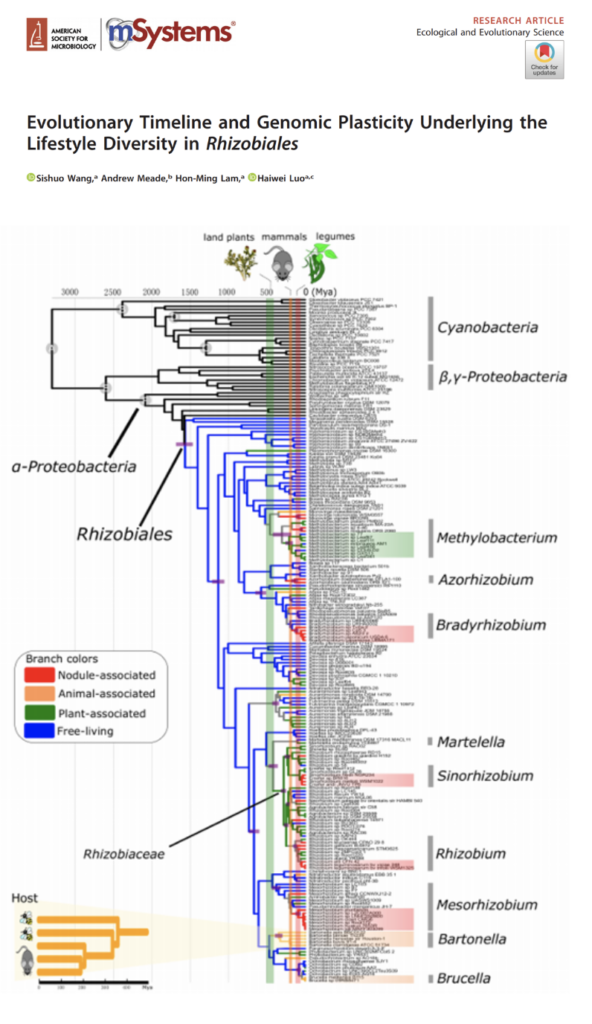
Evolutionary Timeline and Genomic Plasticity Underlying the Lifestyle Diversity in Rhizobiales
Symbiotic nitrogen fixation is agriculturally and environmentally important. Members of the order Rhizobiales including those capable of nitrogen fixation in nodules as well as pathogens of animals and plants. With over 1,000 Rhizobiales genomes, we traced the first nodulating lineage arisen from either Azorhizobium or Bradyrhizobium 150 to 80 Mya, a time range in general concurrent with the emergence of legumes.
Full Version
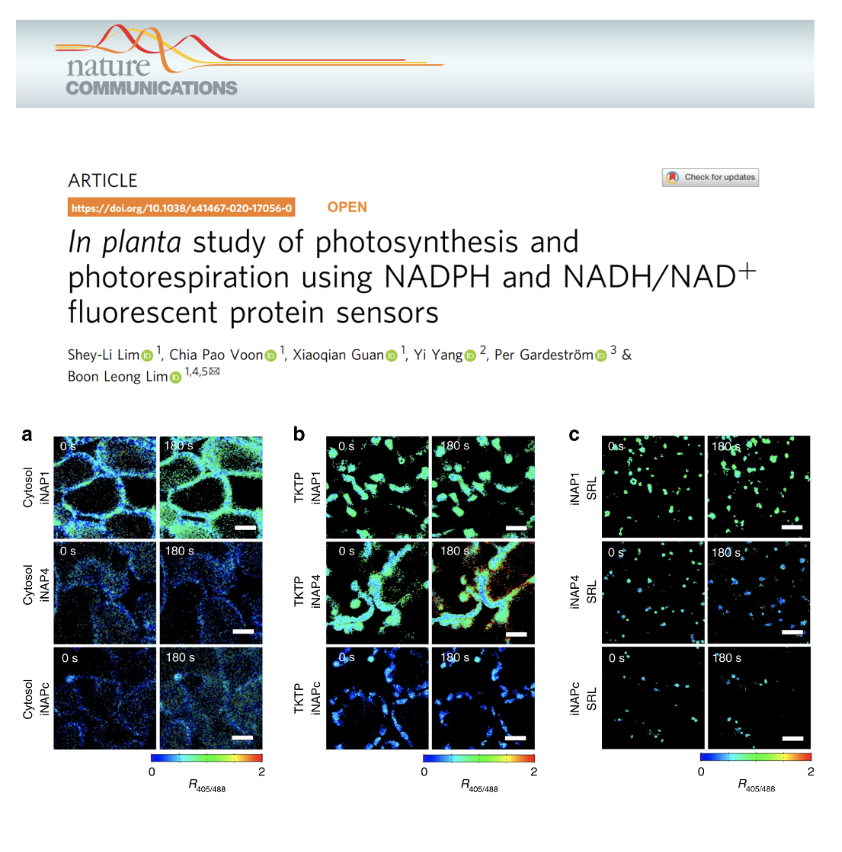
In planta study of photosynthesis and photorespiration using NADPH and NADH/NAD+ fluorescent protein sensors
By employing a NADPH and a NADH/NAD+ sensors, Prof Boon Leong Lim’s team examined in planta dynamic changes in the NADPH pools and NADH/NAD+ ratio in the stroma and cytosol upon illumination.
Full Version
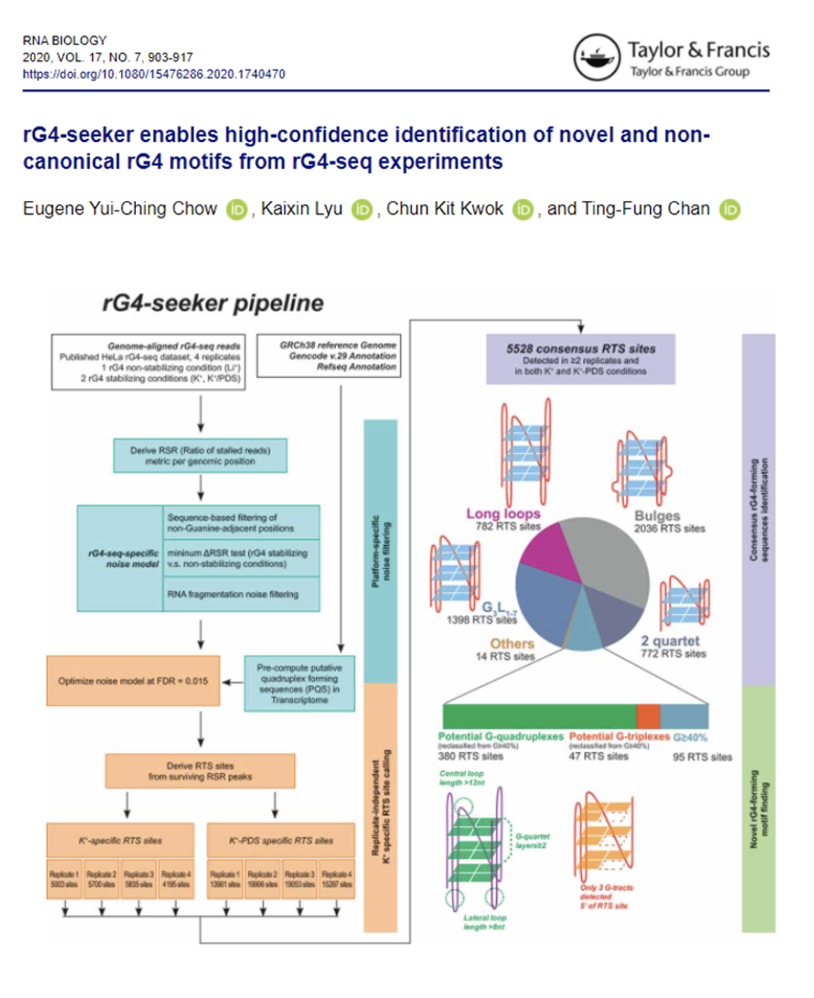
rG4-seeker enables high-confidence identification of novel and non-canonical rG4 motifs from rG4-seq experiments
Team of Prof Ting-Fung Chan has recently developed the rG4-seeker pipeline to detect and map in vitro RNA G-quadruplex (rG4s) structures on a transcriptome-wide scale.
Full Version
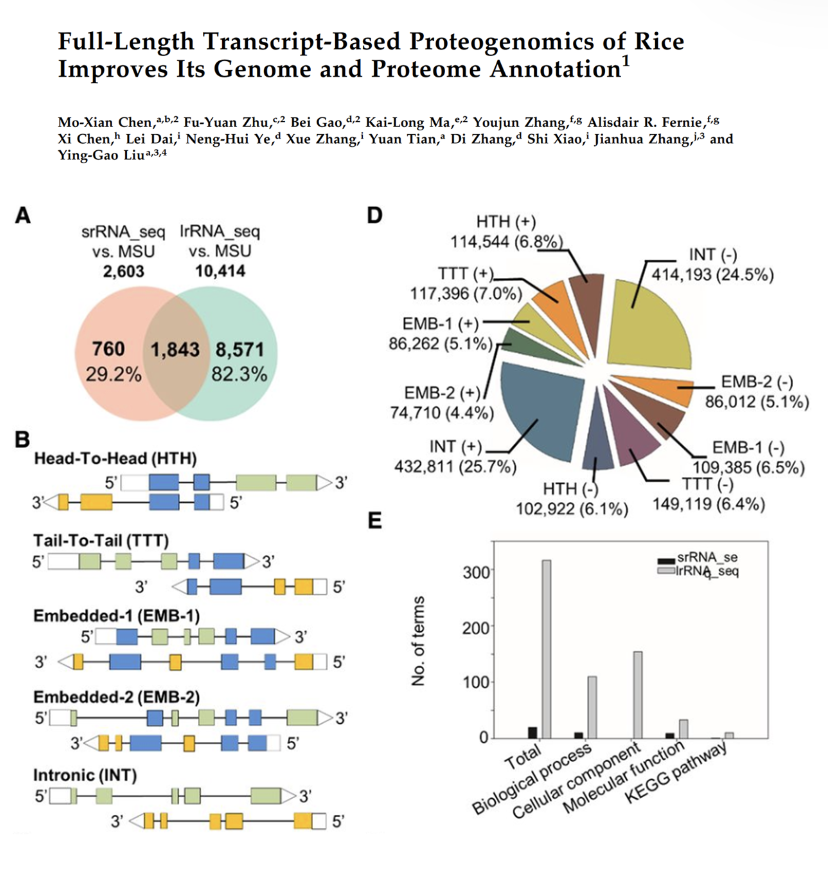
Full-Length Transcript-Based Proteogenomics of Rice Improves Its Genome and Proteome Annotation
Prof Jianhua Zhang, in collaboration with Prof Ying-Gao Liu from Shandong Agricultural University, applied single-molecule long-read RNA sequencing-based proteogenomics to reveal the complexity of the rice transcriptome and its coding abilities.
Full Version
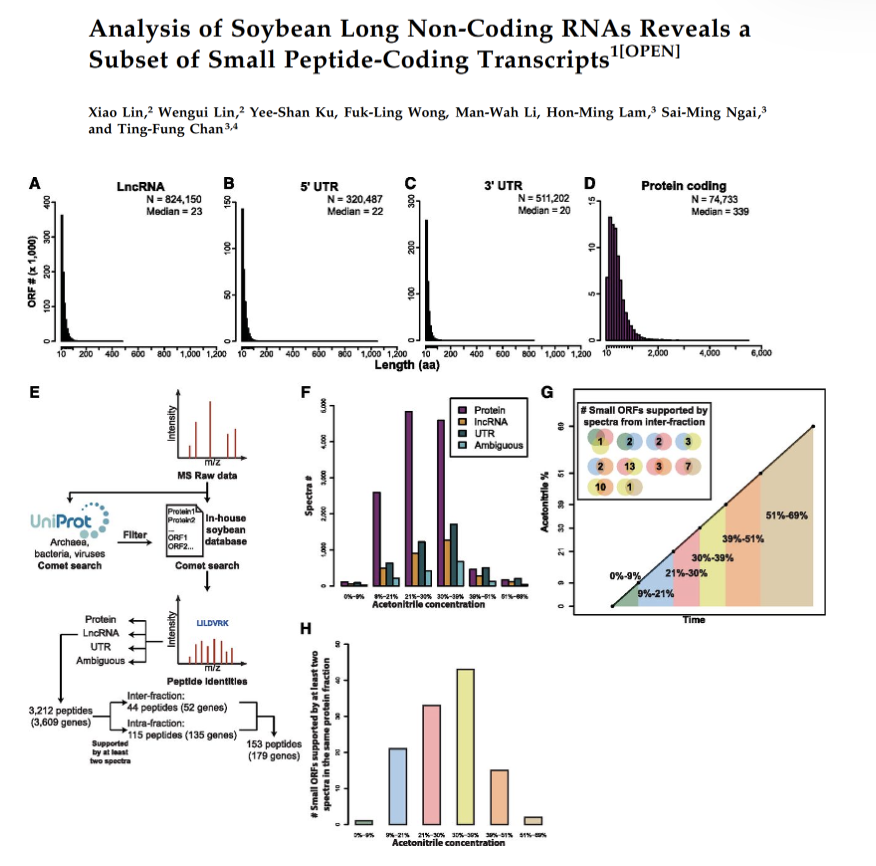
Analysis of Soybean Long Non-Coding RNAs Reveals a Subset of Small Peptide-Coding Transcripts
A total of 332 RNA-seq data samples were used for lncRNA identification. In total, 46,974 and 35,675 lncRNA gene loci were identified in the G. max and G. soja datasets, some of which are differentially expressed under stress conditions. LC-MS/MS was used to validate the translational landscape of these soybean lncRNAs, which are usually regarded as non-coding by definition, paving the way for investigating their functions.
Full Version
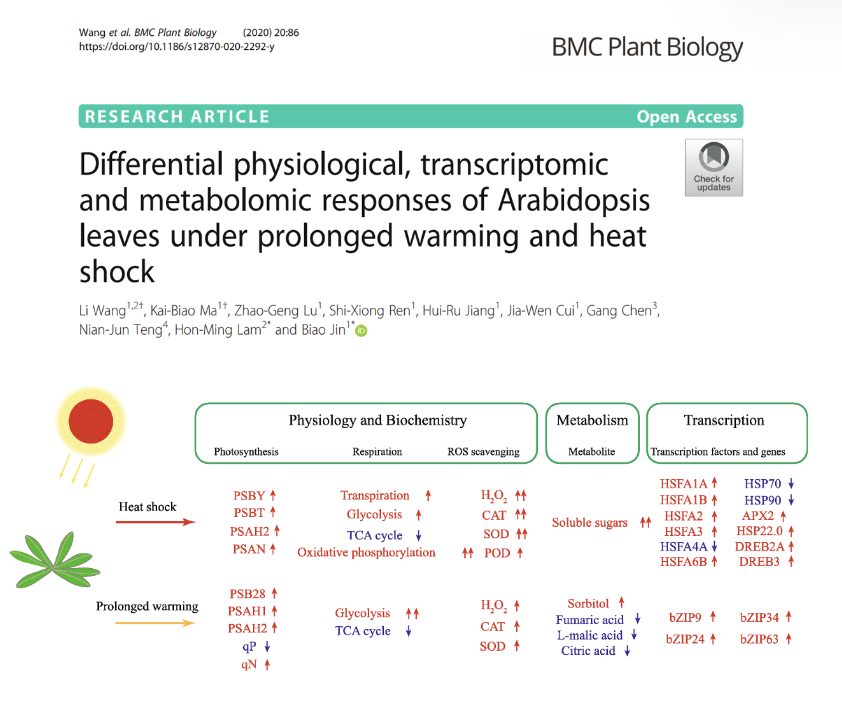
Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock
Elevated temperature as a result of global climate warming, either in form of sudden heatwave (heat shock) or prolonged warming, has profound effects on plants. We performed a comprehensive multi-level comparison of its response toward heat shock and prolonged warming. Physiological, transcriptomic and metabolomic responses of the treated plants are compared. We found that plants respond to heat shock through increasing transpiration rate, the rate of photosynthetic and respiratory electron transfer, the production of ROS, the induction of antioxidant enzymes, and the activation of the HSFA1 pathway. On the other hand, plants response to prolonged warming primarily via decreased stomatal conductance, increased photosynthetic electron transfer rate, inhibited TCA cycle, and activation of a HSFA1-independent response pathway of bZIPs.
Full Version
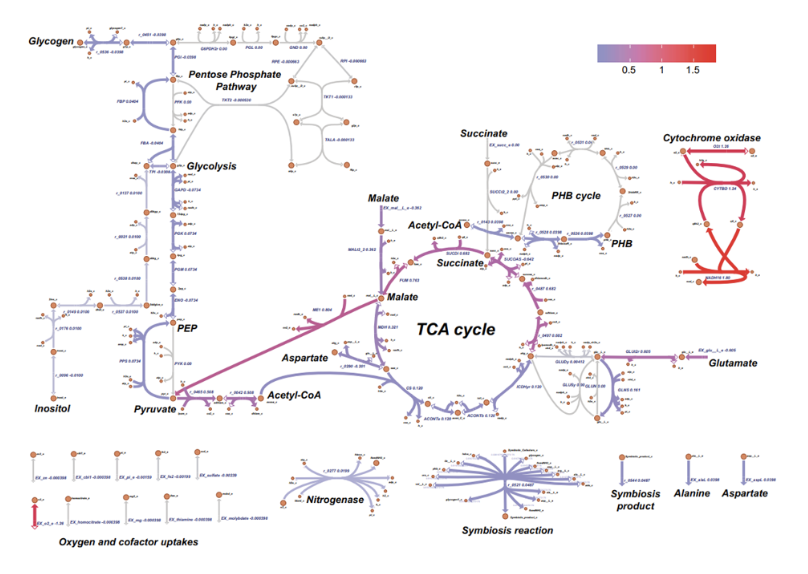
Metabolic Analyses of Nitrogen Fixation in the Soybean Microsymbiont Sinorhizobium fredii Using Constraint-Based Modeling
Further, to understand better the metabolism of the bacteria in nodules, we built a highly complete constraint-based model for S. fredii. Genes controlling the nitrogen fixation process were identified by simulating gene knockouts. Nitrogen-fixing capacities of S. fredii CCBAU45436 in symbiosis with cultivated and wild soybeans were evaluated.
Full Version
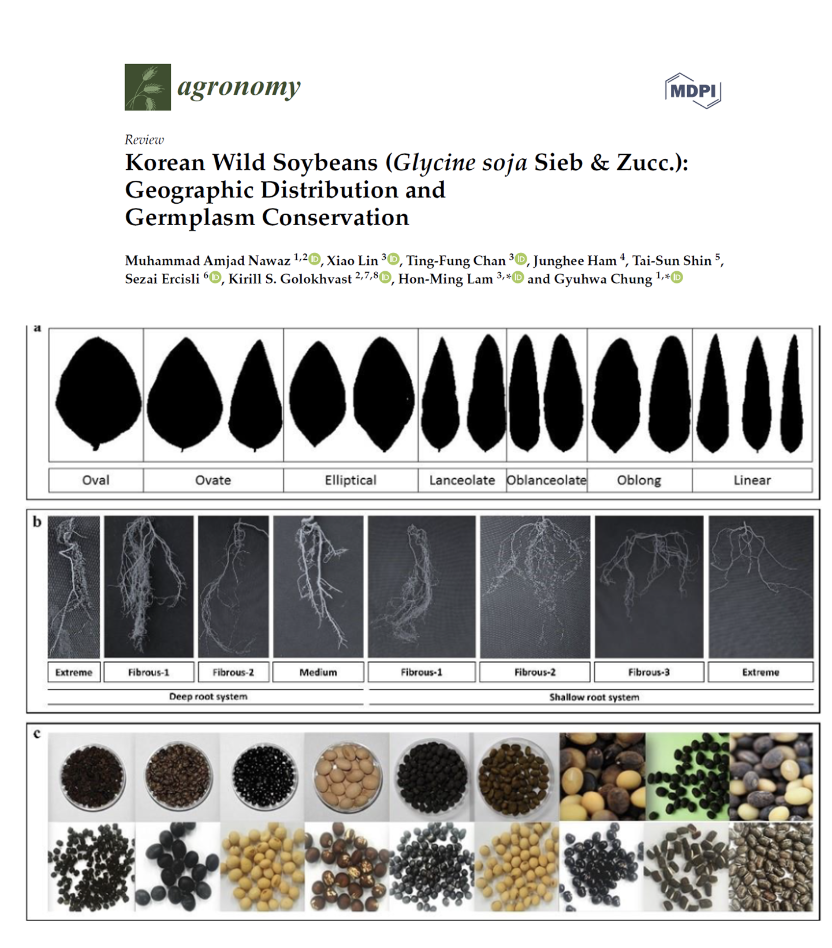
Korean Wild Soybeans (Glycine soja Sieb & Zucc.): Geographic Distribution and Germplasm Conservation
A review on the genetic diversity of Korean wild soybeans
Full Version
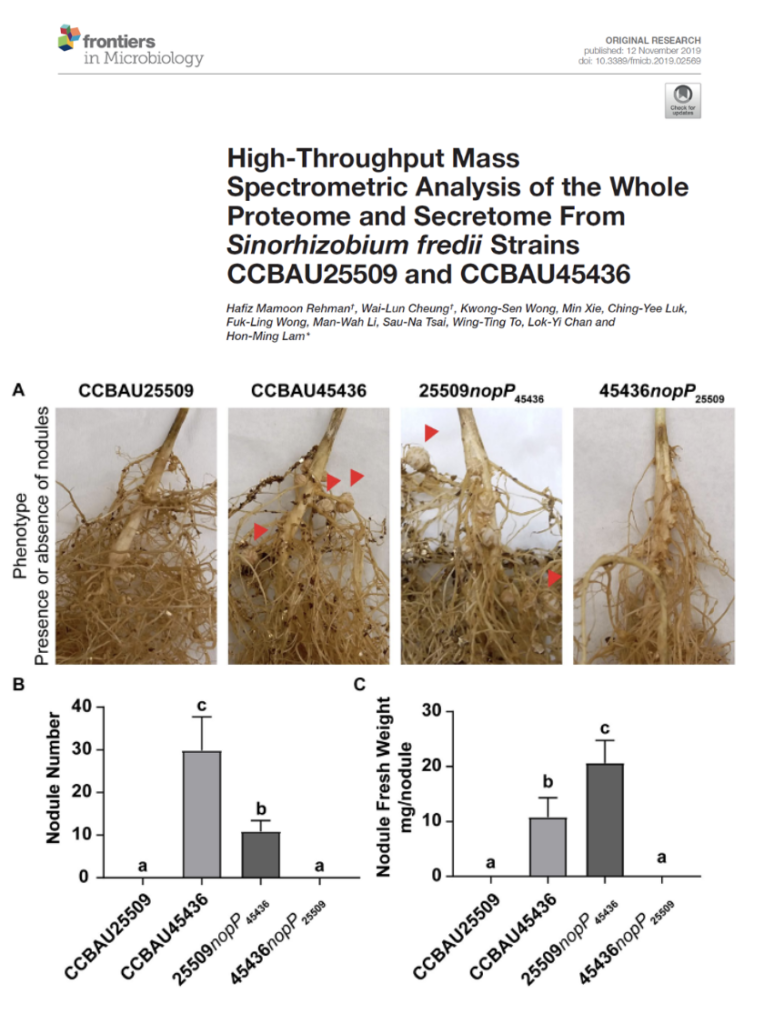
High-Throughput Mass Spectrometric Analysis of the Whole Proteome and Secretome From Sinorhizobium fredii Strains CCBAU25509 and CCBAU45436
We performed comprehensive proteomic analysis of free-living bacteria and bacteroids. The comparison of the secreted proteins between S. fredii, which showed distinct nodulation phenotype in different soybean varieties, have led us to identify NopP as the determinant of host susceptibility in soybean plants.
Full Version

Construction and comparison of three reference-quality genome assemblies for soybean
The AoE team has participated in a joint effort the construction 3 new reference genome assemblies for soybean.
Full Version
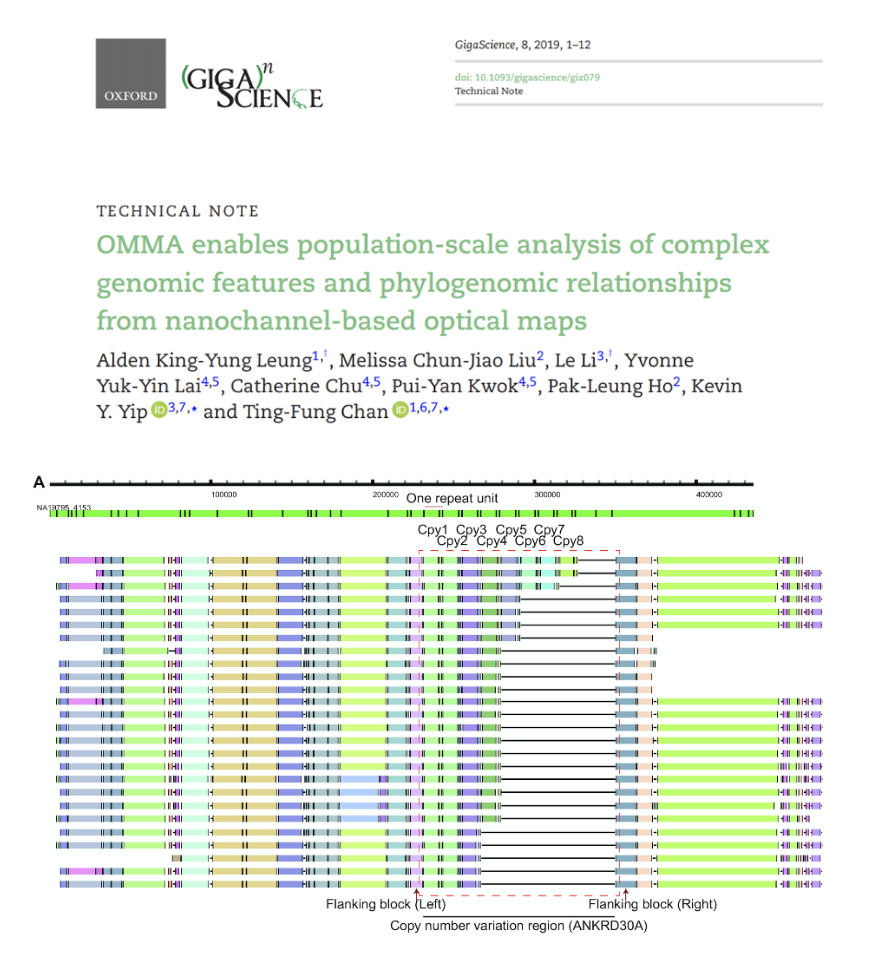
OMMA enables population-scale analysis of complex genomic features and phylogenomic relationships from nanochannel-based optical maps
We have developed a new method, OMMA, that extends optical mapping to the study of complex genomic features by simultaneously interrogating optical maps across many samples in a reference-independent manner. The OMMA software is available at https://github.com/TF-Chan-Lab/OMTools.
Full Version
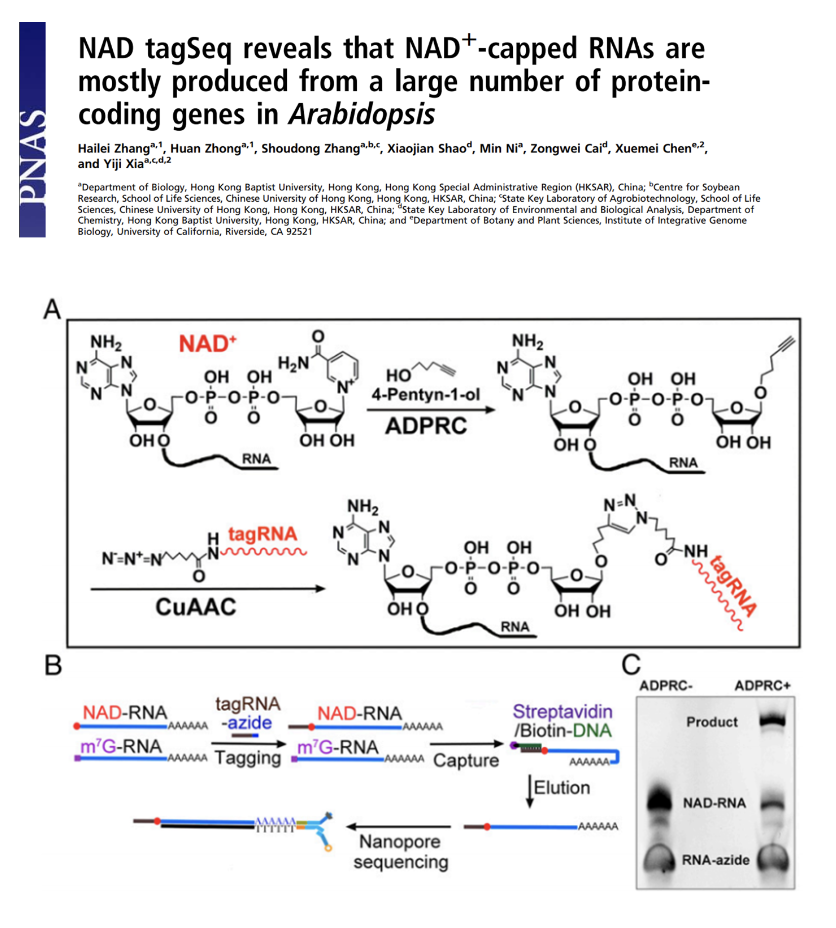
NAD tagSeq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis
Functions of NAD+-capped RNAs are not well explored. A new NAD tagSeq method was developed for transcriptome-wide identification and quantification of NAD+-capped RNAs. The method uses an enzymatic reaction and then a click chemistry reaction to label NAD-RNAs with a synthetic RNA tag. Tagged RNA molecules can be enriched and sequenced for identification and quantification.
Full Version
A reference-grade wild soybean genome
Led by the AoE center, the team has built the first-of-the-world wild soybean reference genome.
Full Version
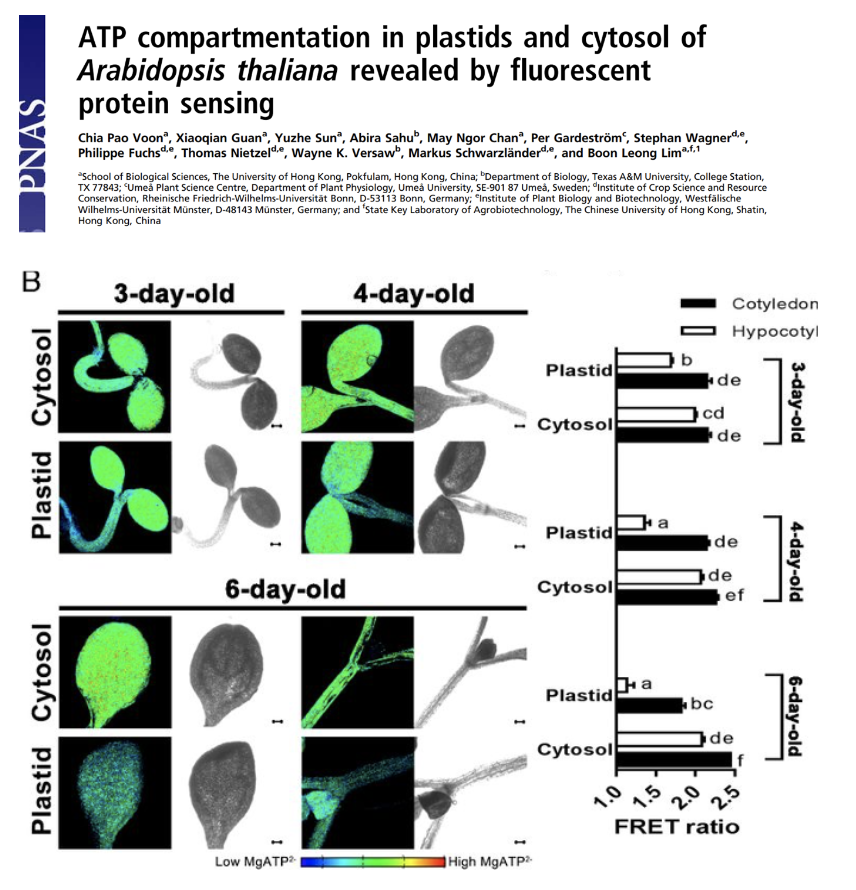
ATP compartmentation in plastids and cytosol of Arabidopsis thaliana revealed by fluorescent protein sensing
By studying in vivo changes of ATP levels in the plastids and cytosol of Arabidopsis thaliana using a FRET-based ATP sensor, Prof Boon Leong Lim’s team showed the differential ATP concentration between plastids and cytosol.
Full Version
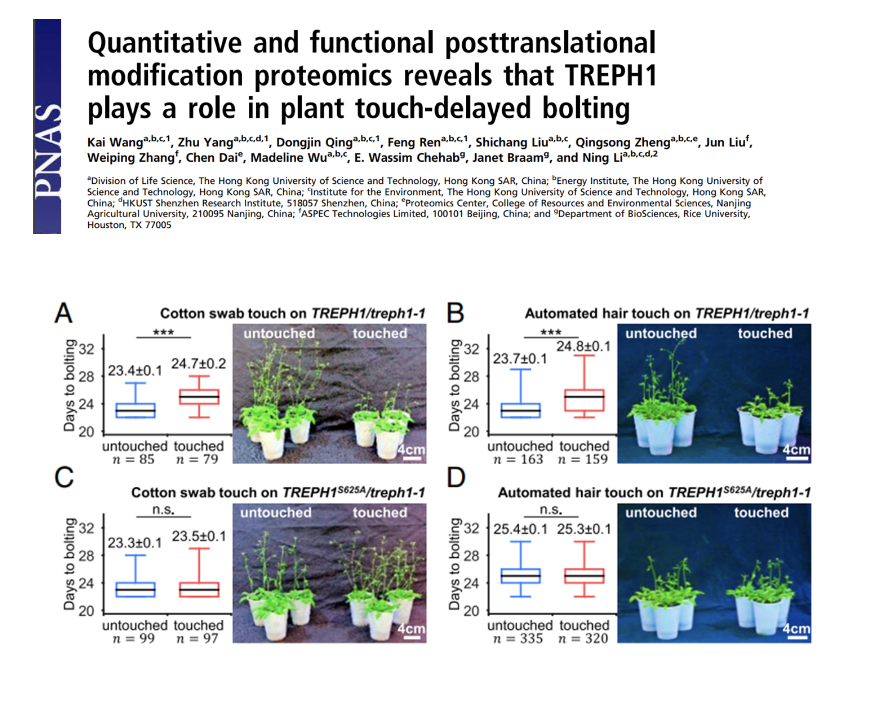
Quantitative and functional posttranslational modification proteomics reveals that TREPH1 plays a role in plant touch-delayed bolting
Environmental mechanical forces, such as wind and touch, trigger gene-expression regulation and developmental changes called “thigmomorphogenesis,” in plants. Touch-responsive phosphopetides were identified through a high-throughput SILIA-based quantitative phosphoproteomics analysis. A previously uncharacterized protein TREPH1 was identified to be rapidly phosphorylated in touch-stimulated plants. The phosphorylation of TREPH1 was later proven to play a role in touch-induced bolting delay.
Full Version
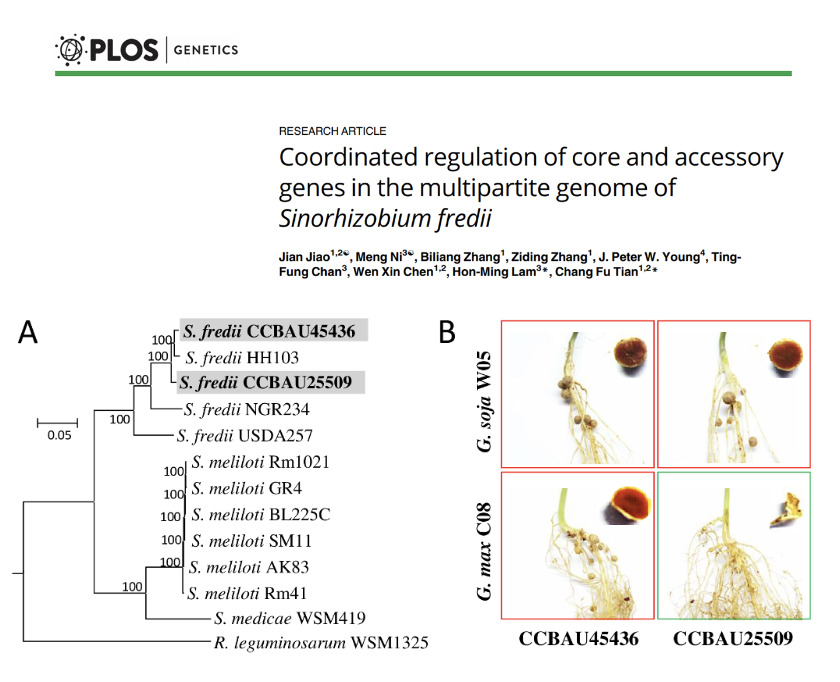
Coordinated regulation of core and accessory genes in the multipartite genome of Sinorhizobium fredii
Sinorhizobium fredii is one of the symbiotic nitrogen-fixing species in the order of Rhizobiales commonly found in Asia, where soybean was originated. We have assembled the complete genomes and carried out transcriptomic study of two S. fredii strains, CCBAU25509 and CCBAU45436, and analyzed them with ten other published genomes of Sinorhizobium to divide their genes into three hierarchical core subsets and accessory genes.
Full Version
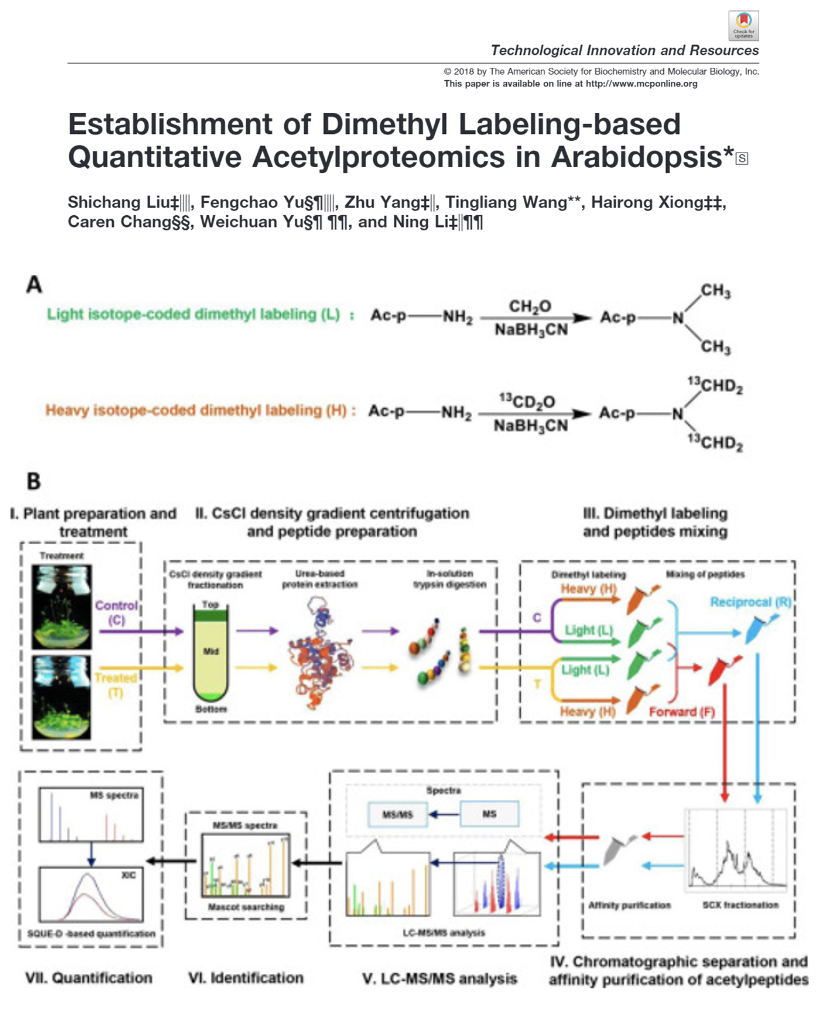
Establishment of Dimethyl Labeling-based Quantitative Acetylproteomics in Arabidopsis
We applied CsCl gradient centrifugation-based protein fractionation, dimethyl labelling-based, mass spectrometry and an in-house software program to carry out quantitative acetylproteomics analysis in Arabidopsis.
Full Version
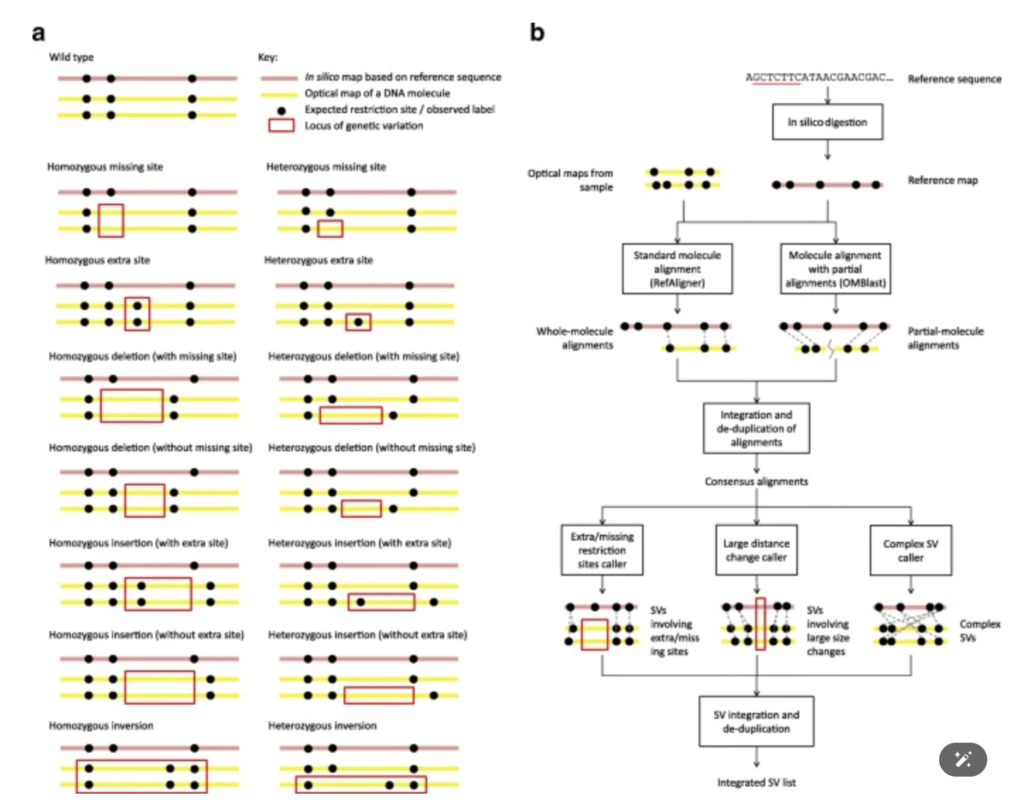
OMSV enables accurate and comprehensive identification of large structural variations from nanochannel-based single-molecule optical maps
We developed a new software OMSV for the identification of structural variations in the genome using optical maps. The OMSV software is available at http://yiplab.cse.cuhk.edu.hk/omsv/.
Full Version

